The website moved to a new address
The lab website is now hosted on a different server. The last update to these pages was in June 2024.
Please visit the new location here.
We gratefully acknowledge funding of our work from various sources including:
 |
 |
 |
||
 |
 |
 |
News
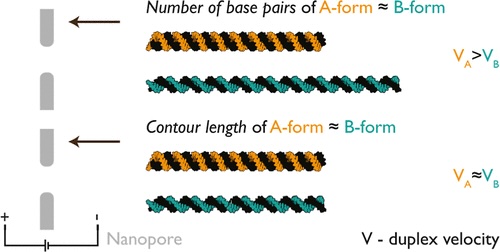
5/6/2024 Forces on RNA:DNA hybrids in nanopores.
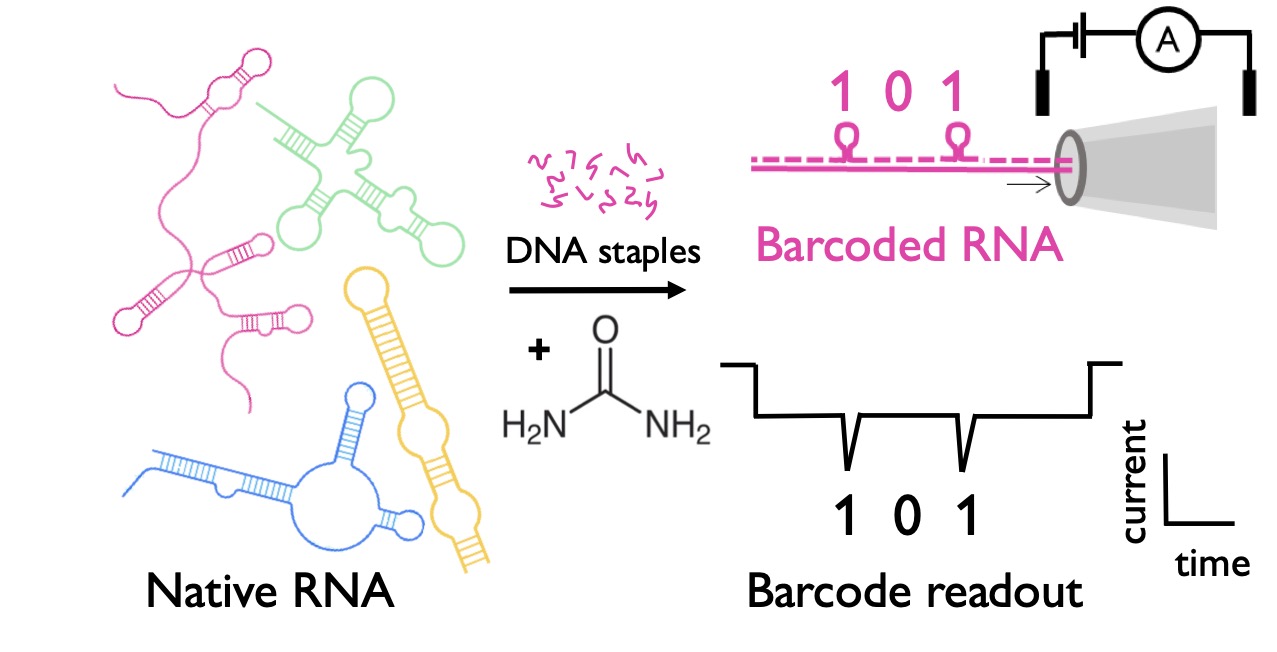
18/5/2024 RNA nanostructure folding (almost) at room temperature .
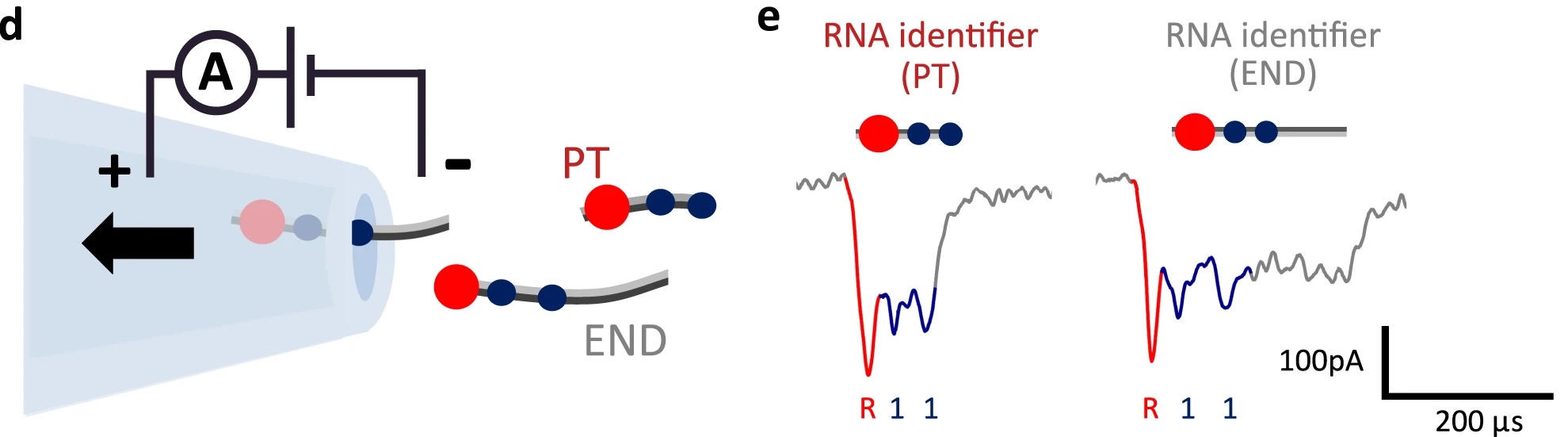
25/2/2024 RNA transcription with solid-state nanopores.
15/10/2023 Michealmas Update.
We welcome Michaelmas term and Siong Chen, who started his MPhil on RNA analysis with nanopores.
We also welcome two visiting PhD students, Sara de Braganca from the Moreno Lab in Madrid and Kevin Neis from the Kjems Lab at Aarhus.
Ran Tivony started his own lab now at Ben-Gurion University. We wish him all the best for his start in these challenging times.
In collaboration with the lab of Gideon Coster we investigated the influence of DNA structures on DNA replication. The paper was published in The Embo Journal.
7/8/2023 dCas9 screened with nanopores.
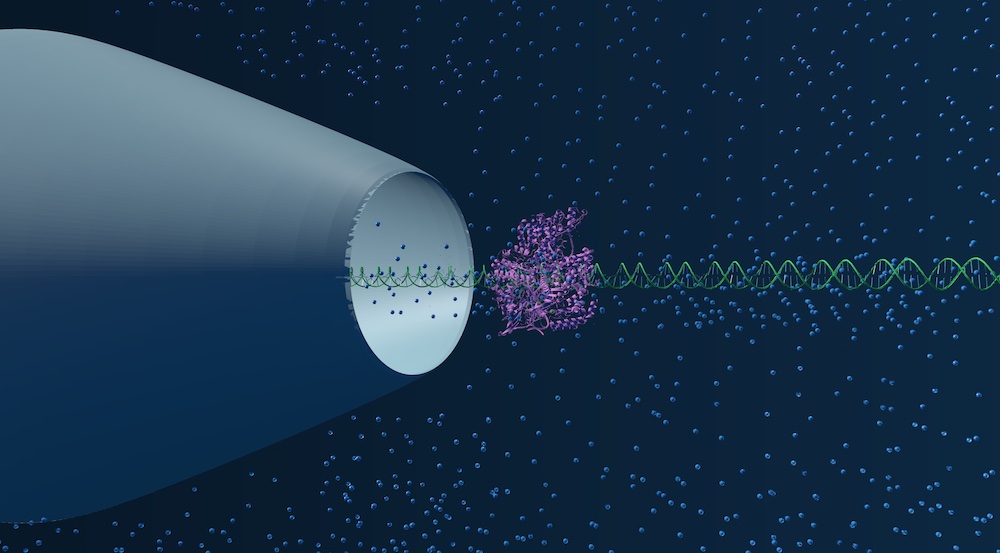 Sarah - in collaboration with Richard Gutierrez from ONT - investigates the binding efficiency of dCas9 to DNA sequences. In her nanopore 'tour de force' Sarah used designed DNA nanostructures for screening dCas9 target sequences with single basepair resolution. Her paper just appeared in Nature Biomed. Eng. Congratulations to Sarah for the great work and excellent collaboration with ONT!
Sarah - in collaboration with Richard Gutierrez from ONT - investigates the binding efficiency of dCas9 to DNA sequences. In her nanopore 'tour de force' Sarah used designed DNA nanostructures for screening dCas9 target sequences with single basepair resolution. Her paper just appeared in Nature Biomed. Eng. Congratulations to Sarah for the great work and excellent collaboration with ONT!
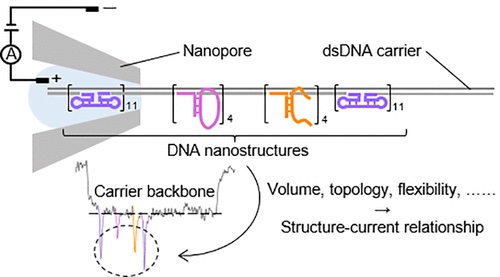
26/7/2023 DNA signals depend on everything.
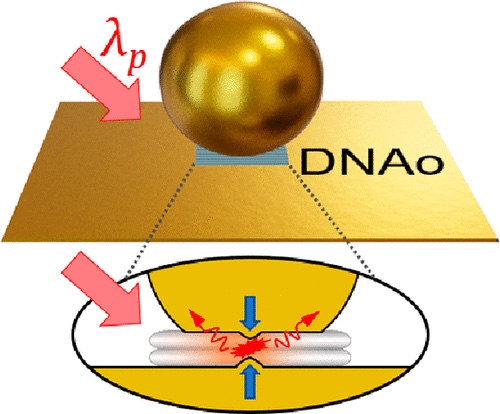
5/7/2023 DNAo nanocavities.