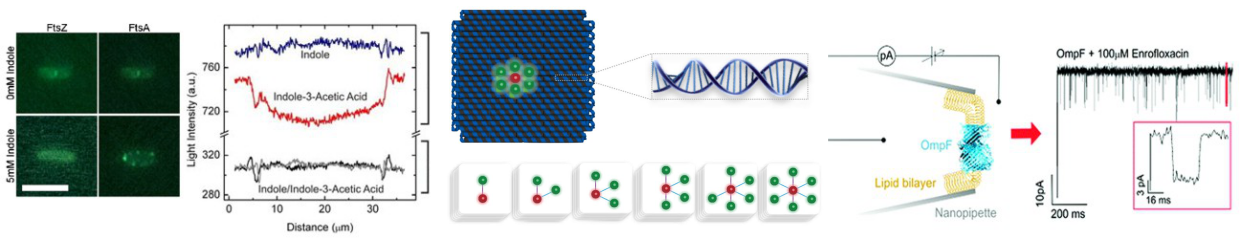
Part III project: Translocation of magnetic colloidal model polymers
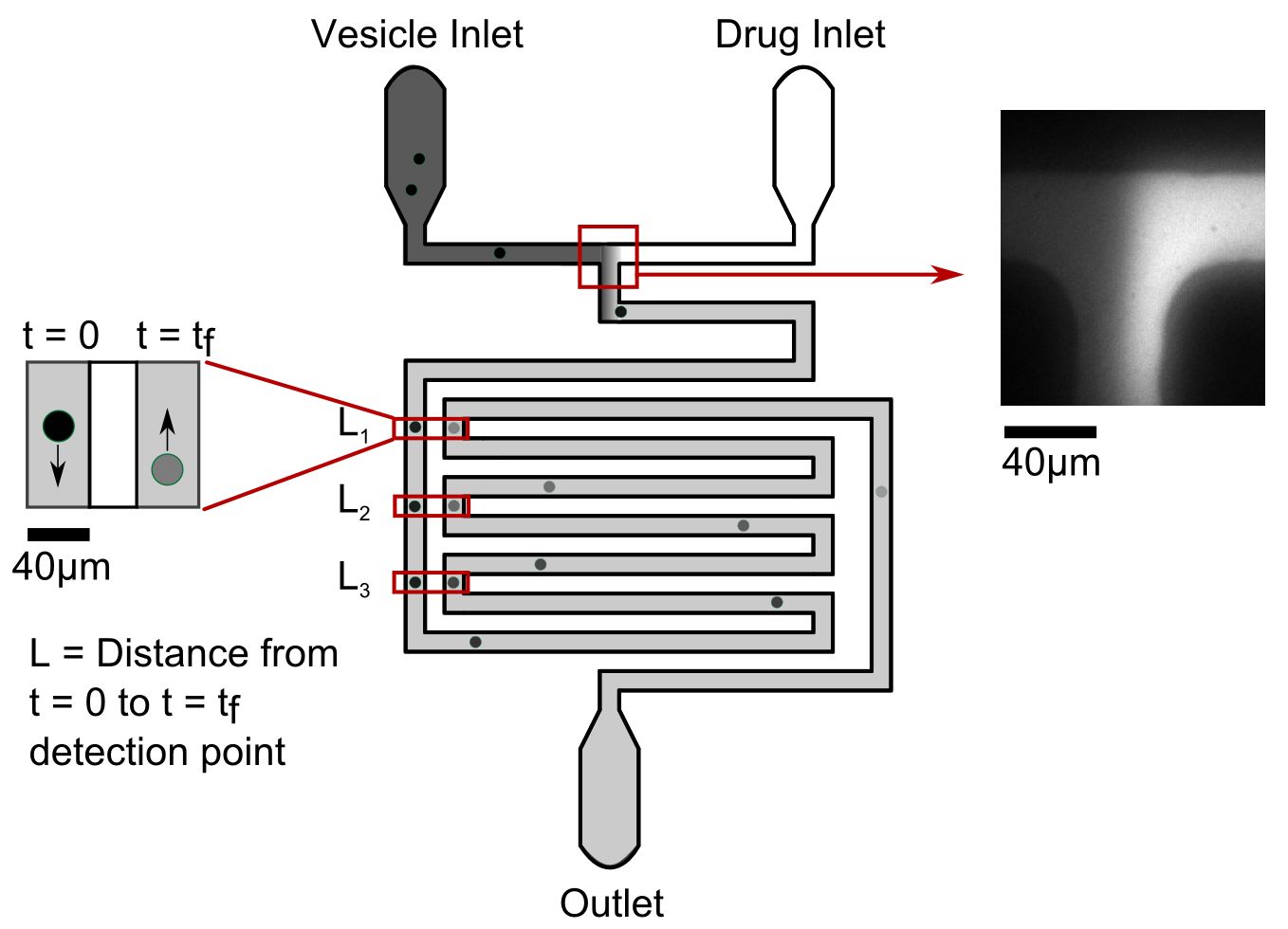
The efficient transport of particles through narrow channels is essential to the functioning of a diverse range of systems, from the biological, where pores control translocation across cell membranes, to the industrial, where porous materials play key roles in filtration, batteries and sensing technologies. Especially complex is the case of polymer translocation, where the chain-like structure of the macromolecule leads to a complex interplay of effects, and as such this process remains poorly understood.
In this project the student will investigate the transport of colloidal model polymers in microfluidic channels. These polymers will be formed from superparamagnetic colloidal spheres that upon the application of an external magnetic field form chains2, previously used as primitive models for polymers2. These experiments will build upon extensive work considering the dynamics of single particles in microfluidic channels3,4, with direct comparison to these results allowing for the impact of directional interparticle interactions upon translocation dynamics to be fully elucidated. By sensitively tuning the system parameters, the effect on translocation of the complex interplay between chain length, channel dimensions, chain stiffness and external driving force can also be studied. Results from this novel model system will be compared to theoretical models for polymer transport, allowing for truly quantitative tests of these models. This will provide unprecedented insight into the fundamental principles governing transport of macromolecules, relevant to the future design of molecular sensors, batteries and selective membranes
Involved researchers:
Alice, Ulrich
Key references:
Deterministic aggregation kinetics of superparamagnetic colloidal particles P. Reynolds, K. E. Klop, F. A. Lavergne, S. M. Morrow, D. G. A. L. Aarts, R. P. A. Dullens, J. Chem. Phys. 143, 214903 (2015)
Fluctuation dynamics of a single magnetic chain R. Silva, R. Bond, F. Plouraboue and D. Wirtz, Phys. Rev. E, 54, 5502, (1996)
Anisotropic diffusion of spherical particles in closely confining microchannels S. L. Dettmer, S. Pagliara, K. Misiunas, U. F. Keyser, Phys. Rev. E. 89, 062305 (2014)
Nondecaying Hydrodynamic Interactions along Narrow Channels K. Misiunas, S. Pagliara, E. Lauga, J. R. Lister, U. F. Keyser, Phys. Rev. Lett. 115, 038301 (2015)
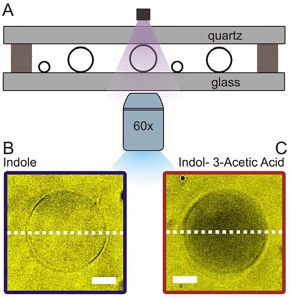
Part III project: High-throughput microfluidics for testing lipid phases
Phospholipid vesicles, also known as liposomes, are a powerful tool and have been used in many scientific disciplines ranging from biology to physics for many years. Nowadays, liposomes find more and more industry applications especially in medicine, pharmacology and biotechnology [1]. Liposomes are an ideal model for a cell's membrane, as it also consists of a phospholipid bilayer. In our experiments, we are using a novel microfluidic method to generate lipid vesicles on a chip to study drug transport processes across the lipid membrane. Our goal is to gain a deeper understanding of the laws that govern cellular drug uptake [2-5]. Cell membranes are complex systems which consist of numerous different types of phospholipids and proteins. In order to mimic a natural cell membrane, we need to successfully create bi- or triphasic liposomes. In this project, you will perform microfluidic experiments to create liposomes with different lipid mixtures and generate a phase diagram. Furthermore, we can perform drug transport experiments on the liposomes to investigate the effect of different phospholipid types on drug permeability of the membrane.Involved researchers:
Jehangir, Kareem, Michael, Ran, Ulrich
Collaborators:
Aleksei Aksimentiev (Urbana-Champaign), Tim Liedl (LMU Munich), Fernando Moreno-Herrero (CSIC Madrid), Oxford Nanopore Technologies
Key references:
[1] Akbarzadeh et al., Nanoscale Research Letters, 2013, 8:102
[2] Deshpande et al., Nature Communications, 2016, 7:10447
[3] Cama et al., Lab Chip, 2014, 14:2303
[4] Cama et al., Scientific Reports, 2016, 6:32824
[5] Al Nahas et al. LabChip, 2018, under review
Driven fluid jets in hybrid DNA origami and graphene nanopores
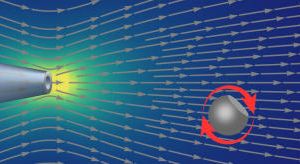
Fluid jets are found in nature at all length scales - microscopic to cosmological. In this project the student will investigate electroosmotically driven, submerged fluidic jets emerging from single glass nanochannels capped with DNA origami structures or graphene. Hybrid pore systems are of interest for improved bio sensing applications and for engineering of model biological systems - and are currently poorly understood at a fundamental level. The student will learn how to produce glass nanopores and assemble hybrid nanopore systems. The glass nanopores have diameters ranging from 15 to 150 nm. Our recently developed optical tweezers based technique allows one to measure minute flows with flow rates of a few pL/s. Our novel technique will be used to map out the 3 dimensional velocity fields of the fluid emerging from the nanochannel. This work builds upon extensive work studying electroosmotically driven flows out of bare glass nanopores. The student will compare the measurements with classical solutions of the Navier-Stokes equation, e.g. the Landau-Squire solution for a point jet. The experiments test the limits of the continuum models - Poisson-Boltzmann and Stokes equations - at nanometre lengths scales by using different types of ions in aqueous solution. The results enhance our understanding of fluidic flows for biosensing, water filtration and osmotic energy production and creation of model systems to study biological processes.Involved researchers:
Jeff, Kurt, Alice, Ulrich
Key references:
N. Laohakunakorn, B. Gollnick, F. Moreno-Herreo, D. G. A. L. Aart, R. P. A Dullens, S. Ghosal, and U. F. Keyser. A Landau-Squire nanojet. Nano Letters, 3(11):5141-5146, 2013.
N. Laohakunakorn and U. F. Keyser. Electroosmotic flow rectification in conical nanopores. Nanotechnology, 26(27):275202, 2015
N. Laohakunakorn, V. V. Thacker, M. Muthukumar, and U. F. Keyser. Electroosmotic flow reversal outside glass nanopores. Nano Letters, 15(1):695-702, 2014.
G. Rempfer, S. Ehrhardt, N. Laohakunakorn, G. B. Davies, U. F. Keyser, C. Holm, and J. de Graff. Selective Trapping of DNA using Glass Microcapillaries. Langmuir, 32(33):8525-8532, 2016.
Part III project: Physics of Molecular Transport through Graphene Nanopores
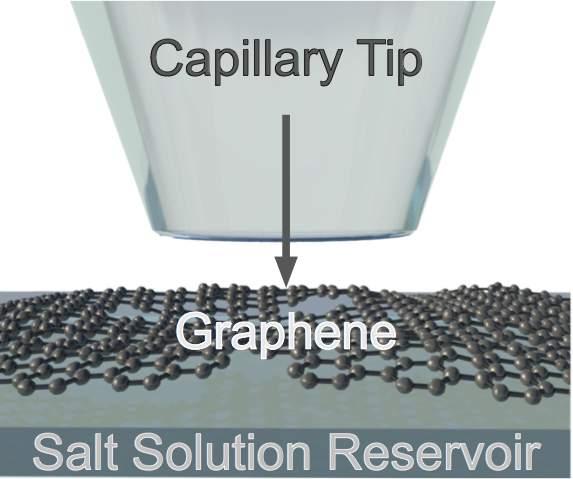
There has probably never existed a material that has been assigned as many superlatives, and garnered as much hype as graphene. By exploiting the 'strongest' and 'thinnest' material, this project aims to eventually achieve high resolution sensing across the single atomic plane of graphene analysing DNA structures and digital barcodes [1]. The experimental setup features a novel method of contacting the graphene membrane; glass capillaries are brought into contact with the membrane [a] until a seal is established [b]. This graphene tipped capillary is then exposed to a variety of electrolytes or materials allowing for electrical characterisation and ionic data collection.
Defects within the membrane have shown selective porosity toward cations [2,3]. Currently, work is being
made to tune this selectivity toward anions. Studying the effect of the type of salt and solvent used towards
this effort is of particular interest. Understanding these extrinsic effects would enhance the single molecule
sensing in this novel system.
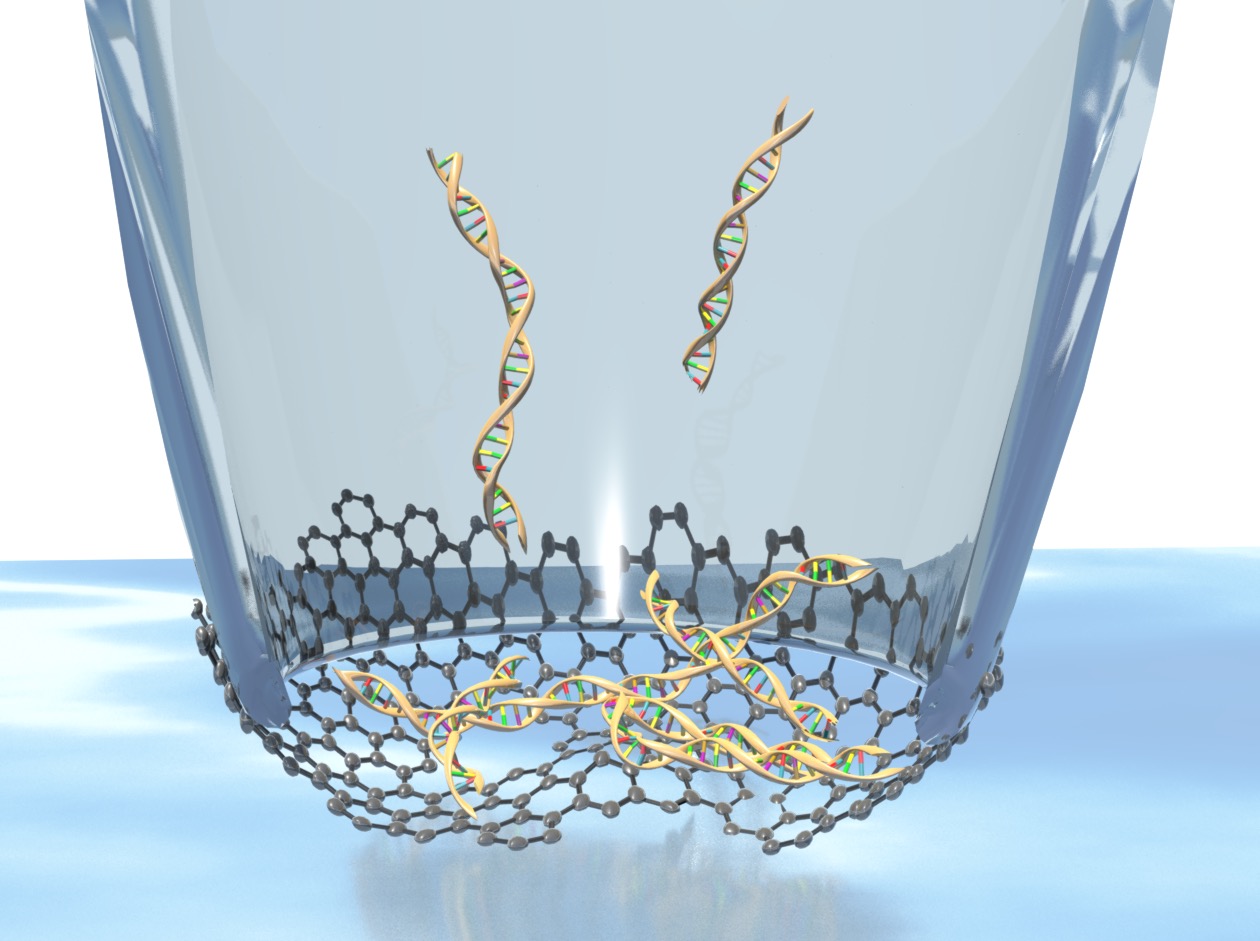
In addition, methods are currently being used to create 3-10nm diameter pores within the membrane via
dielectric breakdown [4]. These larger pores are used to sense the translocation of DNA across the
membrane [c]. This very exciting research comes with challenges in choices of solvent, consideration of
DNA-graphene interactions and electrical noise.
Involved researchers:
Mustafa, Jeff, Ulrich
Key references:
[1] Bell, Nicholas AW, and Ulrich F. Keyser. "Digitally encoded DNA nanostructures for multiplexed, single-molecule
protein sensing with nanopores." (2016)
[2] Walker, Michael I., et al. "Measuring the proton selectivity of graphene membranes." Applied Physics Letters 107.21
(2015): 213104
[3] Walker, Michael I., et al. "Extrinsic Cation Selectivity of 2D Membranes." ACS nano 11.2 (2017): 1340-1346
[4] Kuan, Aaron T., et al. "Electrical pulse fabrication of graphene nanopores in electrolyte solution." Applied physics
letters 106.20 (2015): 203109